To determine if a subhalo has undergone a major merger, I am attempting to measure the mass ratio using the MergerTree.
Then I found out about numMergers function in Sublink, and I have a question about how it works.
When I referred to this discussion forum( Major mergers identifications ), I could see that this function was as follows.
" There, a particular kind of mass ratio (defined as the maximum mass of some type, for both objects) is computed. "
When examining the definitions of fpMass and npMass, it appears that the stellar mass of each object is defined as the value at the time of its maximum. I don't understand why it calculates the mass ratio with the stellar mass at different times. In the Sublink , Rodriguez-Gomez+ (2015) that I referred to, it seems that fpMass uses the stellar mass of the FirstProgenitor at the same time(t_max of nextProgenitor) as when the mass of the nextProgenitor is at its maximum. Is there a reason why we don't use it this way?
The following is the result obtained by running the modified version of the existing code. In addition to the maximum mass returned by maxPastmass, it now also informs us of the time (Snapshot) when the maximum occurs.
< MaxPastMass >
def maxPastMass2(tree, index, partType='stars'):
""" Get maximum past mass (of the given partType) along the main branch of a subhalo
specified by index within this tree. """
ptNum =4
branchSize = tree['MainLeafProgenitorID'][index] - tree['SubhaloID'][index] + 1
masses = tree['SubhaloMassType'][index: index + branchSize, ptNum]
return np.max(masses), tree['SnapNum'][index: index + branchSize][np.argmax(masses)]
< NumMergs >
numMergers=0
numNp=0
numMid=[]
index=0
minMassRatio=1/10
invMassRatio = 1.0 / minMassRatio
rootID=tree['SubhaloID'][index]
fpID=tree['FirstProgenitorID'][index]
massPartType='stars
while fpID != -1 :
fpIndex= index + (fpID-rootID)
fpMass,fpmax=maxPastMass2(tree,fpIndex,massPartType)
npID= tree['NextProgenitorID'][fpIndex]
numNp=0
while npID!= -1 :
npIndex = index + (npID - rootID)
npMass,npmax=maxPastMass2(tree,npIndex,massPartType)
if fpMass >0.0 and npMass >0.0 :
ratio=npMass/fpMass
if ratio >= minMassRatio and ratio <= invMassRatio:
numMergers += 1
print("#Sn=",tree['SnapNum'][fpIndex])
print("FpMass: ",fpMass,"at SN:",fpmax)
print("NpMass: ",npMass,"at SN:",npmax,"\n")
npID = tree['NextProgenitorID'][npIndex]
fpID = tree['FirstProgenitorID'][fpIndex]
#Sn= 82
FpMass: 5.257069 at SN: 82
NpMass 0.5819944 at SN: 74
#Sn= 58
FpMass: 3.2475514 at SN: 58
NpMass: 0.60512143 at SN: 55
#Sn= 35
FpMass: 0.60444576 at SN: 35
NpMass: 0.4369604 at SN: 35
#Sn= 23
FpMass: 0.0460811 at SN: 23
NpMass: 0.0068318914 at SN: 22
#Sn= 19
FpMass: 0.008799356 at SN: 19
NpMass: 0.0015845126 at SN: 18
#Sn= 3
FpMass: 3.4521076e-05 at SN: 3
NpMass: 5.304361e-06 at SN: 2
As seen in the above results, I can observe that the snapshots of fnMass and npMass used are different from each other.
I'll be grateful for your help .
Dylan Nelson
5 Apr '23
Hi Hyunmi,
You are correct, the numMergers() example code is just meant as an example, and it does not implement exactly the procedure described in Rodriguez-Gomez+15. I also agree that a better approach is to take the peak masses at the same time. I found an old email thread on this topic and paste it below.
Hi all,
I have been identifying interacting pairs within IllustrisTNG. Last week, I realized I could modify the illustris_python.sublink.numMergers() code to give me the information I needed. While I was modifying this piece of code, I noticed that there was a small error in the definition of the first progenitor's mass. I'm not sure how impactful this catch will be in the long run, but Lars thought I should share this with you all.
According to the Rodriguez-Gomez+2015 paper, the mass ratio is "based on the stellar masses of the two progenitors taken at t_max, i.e., at the time when the secondary progenitor reaches its maximum stellar mass." The way the code was originally written, the mass ratio is taken as the ratio of the maximum mass of the first progenitor to the maximum mass of the second progenitor. More often than not, these occur at different times. The correct way to do this would be to determine when the second progenitor has its maximum mass, and then determine the first progenitor's mass at that time to find the mass ratio.
I wrote a quick patch to the code (attached) that fixes the problem. To see exactly how different the two codes are, I ran the tree walk-through example from http://www.illustris-project.org/data/docs/scripts/ for the IllustrisTNG z=0 box.
For the 100-105th GroupFirstSub's, the original code gives:
for i in range(100, 105):
tree = il.sublink.loadTree(basePath,99,GroupFirstSub[i],fields=fields)
numMergers = il.sublink.numMergers(tree,minMassRatio=ratio)
print GroupFirstSub[i], numMergers
....:
264412 6
265267 6
265926 4
266559 6
267323 4
My code, run on the same set of galaxies, and using the same minimum mass ratio gives:
for i in range(100, 105):
tree = sublink_new.loadTree(basePath,99,GroupFirstSub[i],fields=fields)
numMergers = sublink_new.numMergers(tree,minMassRatio=ratio)
print GroupFirstSub[i], numMergers
....:
264412 8
265267 8
265926 13
266559 10
267323 6
To get an idea of how this works for a larger sample of galaxies, I also looked at the number of mergers that each version of the code found as a function of group mass for the first 10,000 GroupFirstSub galaxies. There doesn't seem to be a strong dependence on group mass, but this definitely shows that the original illustris_python.sublink.numMergers() code significantly underestimates the number of mergers that occur above a given mass ratio.
I'm not sure if this error is also in the IDL or Matlab versions of the code, but I suspect that it might be. I hope some of you find this useful in the future.
Cheers,
Kelly Blumenthal
Updated sublink.py file:
def maxPastMass(tree, index, partType='stars'):
""" Get maximum past mass (of the given partType) along the main branch of a subhalo
specified by index within this tree. """
ptNum = il.util.partTypeNum(partType)
branchSize = tree['MainLeafProgenitorID'][index] - tree['SubhaloID'][index] + 1
masses = tree['SubhaloMassType'][index: index + branchSize, ptNum]
#added:
snaps = tree['SnapNum'][index: index + branchSize]
return np.max(masses), snaps[np.where(masses == np.max(masses))[0]]
#added:
def massAtSnap(tree, index, snapnum, partType='stars'):
""" Get mass (of the given partType) along the main branch of a subhalo
at a specific snap shot. """
ptNum = il.util.partTypeNum(partType)
branchSize = tree['MainLeafProgenitorID'][index] - tree['SubhaloID'][index] + 1
masses = tree['SubhaloMassType'][index: index + branchSize, ptNum]
snaps = tree['SnapNum'][index: index + branchSize]
idx = np.where(snaps == snapnum)[0]
return masses[idx]
def numMergers(tree, minMassRatio=1e-10, massPartType='stars', index=0):
""" Calculate the number of mergers in this sub-tree (optionally above some mass ratio threshold). """
# verify the input sub-tree has the required fields
reqFields = ['SubhaloID', 'NextProgenitorID', 'MainLeafProgenitorID',
'FirstProgenitorID', 'SubhaloMassType', 'SnapNum']
if not set(reqFields).issubset(tree.keys()):
raise Exception('Error: Input tree needs to have loaded fields: '+', '.join(reqFields))
numMergers = 0
invMassRatio = 1.0 / minMassRatio
# walk back main progenitor branch
rootID = tree['SubhaloID'][index]
fpID = tree['FirstProgenitorID'][index]
while fpID != -1:
fpIndex = index + (fpID - rootID)
#removed: fpMass = maxPastMass(tree, fpIndex, massPartType)
# explore breadth
npID = tree['NextProgenitorID'][fpIndex]
while npID != -1:
npIndex = index + (npID - rootID)
#removed: npMass = maxPastMass(tree, npIndex, massPartType)
#added:
npMass, npSnap = maxPastMass(tree, npIndex, massPartType)
# count if both masses are non-zero, and ratio exceeds threshold
#removed: if fpMass > 0.0 and npMass > 0.0:
#added:
if npMass > 0.0:
#added:
fpMass = massAtSnap(tree, fpIndex, npSnap, massPartType)
if fpMass > 0.0:
ratio = npMass / fpMass
if ratio >= minMassRatio and ratio <= invMassRatio:
numMergers += 1
npID = tree['NextProgenitorID'][npIndex]
fpID = tree['FirstProgenitorID'][fpIndex]
return numMergers
Hyunmi Song
5 Apr '23
Thank you so much for your help! I confirmed that the modified code you provided and the code I modified myself match perfectly.
Thank you again for your kind assistance. It was truly valuable and greatly appreciated
Dear Dylan,
Additionally, I have a question. I would like to create a function similar to numMergers using LhaloTree.
However, it seems that LhaloTree does not have a function like numMergers, is that correct?
If the function doesn't exist, I'm trying to create it myself. But, I'm having difficulty finding the method in LhaloTree to find the next progenitor.
I understand the definition of NextProgenitor in LhaloTree to be as follows :
"The index of the next subhalo from the same snapshot which shares the same descendant, if any (-1 if this is the last). Indexes this TreeX group.".
Does this mean that the value of NextProgenitor refers to the direct index in this dataset when the value is obtained from the entire branch (not just the main branch) through loadTree?
However, when I try to apply this, I encounter the following error :
The way of finding the nextprogenitor in SubLink was as follows :
< In numMergers function >
rootID=tree['SubhaloID'][index]
fpID = tree['FirstProgenitorID'][index]
while fpID != -1:
fpIndex = index + (fpID - rootID)
npID = tree['NextProgenitorID'][fpIndex]
while npID != -1:
npID = tree['NextProgenitorID'][npIndex]
fpID = tree['FirstProgenitorID'][fpIndex]
and I'm sure there must be a similar way of finding it in LhaloTree, but I'm not sure what it is.
Please let me know if there is any issue with the approach I have taken! Thank you as always.
Dylan Nelson
3 May '23
I am not using LHaloTree much, but I think the issue is the difference between these two statements:
(1) "Indexes this TreeX group."
(2) "Does this mean that the value of NextProgenitor refers to the direct index in this dataset when the value is obtained from the entire branch (not just the main branch) through loadTree?"
I would have agreed with you (it would make sense like #2), but it isn't exactly the case. What is written in #1 is correct.
In particular, if you actually look at the LHaloTree files you see many groups named Tree{X}. As the documentation explains, one of these groups can contain subhalos of different halos, and what subhalo is where is a bit confusing. The loadTree() function handles this, and returns to you the MPB or the full tree for that subhalo. However, it isn't immediately obvious what Tree{X} group it comes from.
In your particular case above, this subhalo is inside Tree0 of trees_sf1_099.0.hdf5, and all the datasets of this tree have a size of 218547998. So indeed you will find all your NextProgenitor indices inside the datasets of this tree (153231757 will be in bounds).
In practice, then, you could use the treeOffsets() function to get the file and tree number (the first and third returns, respectively). Then, you would probably want to manually load (with h5py) the datasets of this Tree{X} group, from the given file, and then follow the NextProgenitor links as you have above.
Thắng Huỳnh
8 Feb
Hi Dylan,
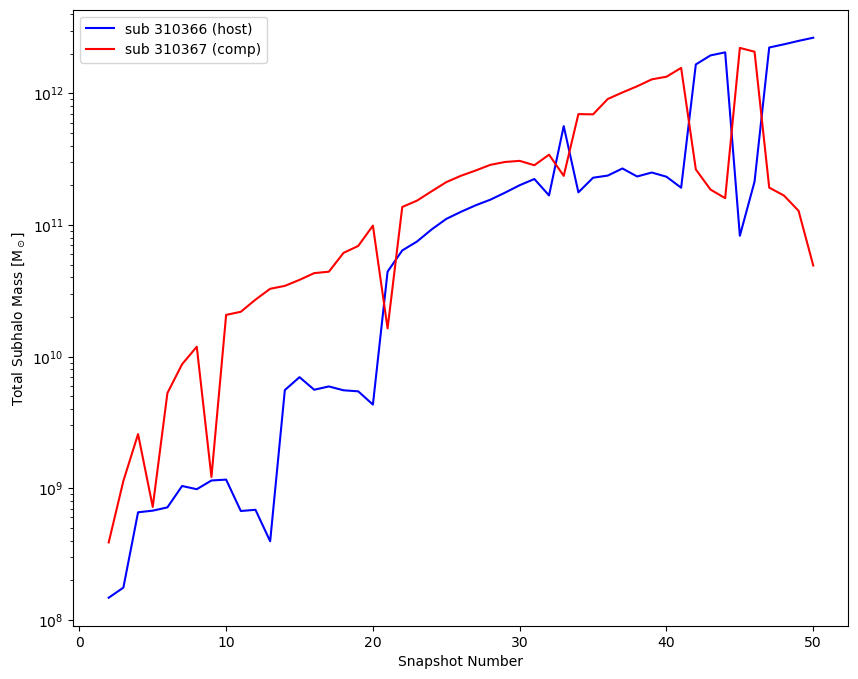
Regarding the mass evolution of subhalos in IllustrisTNG in a merger, I plotted the mass versus time for two interacting subhalos (which merge at snapshot 50), as shown in the attached figure.
According to Figure 3 in Rodriguez-Gomez+ (2015), as you mentioned in this thread, I notice some differences compared to my figure. I wonder if the sharp drops in the subhalo mass history indicate other merging events occurring at those times. Additionally, why does the mass recover immediately after such rises or drops?
Thank you for your assistance.
Dylan Nelson
8 Feb
This is called "switching", and can easily occur during mergers.
The masses recover because they have never actually physically decreased, simply Subfind decided to assign different particles to that subhalo at that particular snapshot.
So is it reasonable to take the mass at t_max (the time when the second progenitor reaches its peak mass) when there is a dip in the first progenitor at that time (according to my plot)?
Dylan Nelson
10 Feb
I think you can see, by eye, reasonable values of the merger mass ratio, given the two lines you have plotted. That is, if you "unswitch" them by eye.
It would be best to come up with a definition that recovers such a value.
Thắng Huỳnh
10 Feb
I do not see how the numMergers function in Sublink deals with "switching." Does this mean it may over/under-estimate some merger ratios?
That is true. It would be hard to get a perfect answer in all cases, as some trees are difficult. Averaging over a sizeable sample, however, would make this effect unimportant.
Hello,
To determine if a subhalo has undergone a major merger, I am attempting to measure the mass ratio using the MergerTree.
Then I found out about numMergers function in Sublink, and I have a question about how it works.
When I referred to this discussion forum( Major mergers identifications ), I could see that this function was as follows.
" There, a particular kind of mass ratio (defined as the maximum mass of some type, for both objects) is computed. "
When examining the definitions of fpMass and npMass, it appears that the stellar mass of each object is defined as the value at the time of its maximum. I don't understand why it calculates the mass ratio with the stellar mass at different times. In the Sublink , Rodriguez-Gomez+ (2015) that I referred to, it seems that fpMass uses the stellar mass of the FirstProgenitor at the same time(t_max of nextProgenitor) as when the mass of the nextProgenitor is at its maximum. Is there a reason why we don't use it this way?
The following is the result obtained by running the modified version of the existing code. In addition to the maximum mass returned by maxPastmass, it now also informs us of the time (Snapshot) when the maximum occurs.
< MaxPastMass >
< NumMergs >
As seen in the above results, I can observe that the snapshots of fnMass and npMass used are different from each other.
I'll be grateful for your help .
Hi Hyunmi,
You are correct, the
numMergers()example code is just meant as an example, and it does not implement exactly the procedure described in Rodriguez-Gomez+15. I also agree that a better approach is to take the peak masses at the same time. I found an old email thread on this topic and paste it below.Hi all,
I have been identifying interacting pairs within IllustrisTNG. Last week, I realized I could modify the
illustris_python.sublink.numMergers()code to give me the information I needed. While I was modifying this piece of code, I noticed that there was a small error in the definition of the first progenitor's mass. I'm not sure how impactful this catch will be in the long run, but Lars thought I should share this with you all.According to the Rodriguez-Gomez+2015 paper, the mass ratio is "based on the stellar masses of the two progenitors taken at t_max, i.e., at the time when the secondary progenitor reaches its maximum stellar mass." The way the code was originally written, the mass ratio is taken as the ratio of the maximum mass of the first progenitor to the maximum mass of the second progenitor. More often than not, these occur at different times. The correct way to do this would be to determine when the second progenitor has its maximum mass, and then determine the first progenitor's mass at that time to find the mass ratio.
I wrote a quick patch to the code (attached) that fixes the problem. To see exactly how different the two codes are, I ran the tree walk-through example from http://www.illustris-project.org/data/docs/scripts/ for the IllustrisTNG z=0 box.
For the 100-105th GroupFirstSub's, the original code gives:
My code, run on the same set of galaxies, and using the same minimum mass ratio gives:
To get an idea of how this works for a larger sample of galaxies, I also looked at the number of mergers that each version of the code found as a function of group mass for the first 10,000 GroupFirstSub galaxies. There doesn't seem to be a strong dependence on group mass, but this definitely shows that the original illustris_python.sublink.numMergers() code significantly underestimates the number of mergers that occur above a given mass ratio.
I'm not sure if this error is also in the IDL or Matlab versions of the code, but I suspect that it might be. I hope some of you find this useful in the future.
Cheers,
Kelly Blumenthal
Updated
sublink.pyfile:Thank you so much for your help! I confirmed that the modified code you provided and the code I modified myself match perfectly.
Thank you again for your kind assistance. It was truly valuable and greatly appreciated
Dear Dylan,
Additionally, I have a question. I would like to create a function similar to numMergers using LhaloTree.
However, it seems that LhaloTree does not have a function like numMergers, is that correct?
If the function doesn't exist, I'm trying to create it myself. But, I'm having difficulty finding the method in LhaloTree to find the next progenitor.
I understand the definition of NextProgenitor in LhaloTree to be as follows :
"The index of the next subhalo from the same snapshot which shares the same descendant, if any (-1 if this is the last). Indexes this TreeX group.".
Does this mean that the value of NextProgenitor refers to the direct index in this dataset when the value is obtained from the entire branch (not just the main branch) through loadTree?
However, when I try to apply this, I encounter the following error :
The way of finding the nextprogenitor in SubLink was as follows :
and I'm sure there must be a similar way of finding it in LhaloTree, but I'm not sure what it is.
Please let me know if there is any issue with the approach I have taken! Thank you as always.
I am not using LHaloTree much, but I think the issue is the difference between these two statements:
(1) "Indexes this TreeX group."
(2) "Does this mean that the value of NextProgenitor refers to the direct index in this dataset when the value is obtained from the entire branch (not just the main branch) through loadTree?"
I would have agreed with you (it would make sense like #2), but it isn't exactly the case. What is written in #1 is correct.
In particular, if you actually look at the LHaloTree files you see many groups named
Tree{X}. As the documentation explains, one of these groups can contain subhalos of different halos, and what subhalo is where is a bit confusing. TheloadTree()function handles this, and returns to you the MPB or the full tree for that subhalo. However, it isn't immediately obvious whatTree{X}group it comes from.In your particular case above, this subhalo is inside
Tree0oftrees_sf1_099.0.hdf5, and all the datasets of this tree have a size of218547998. So indeed you will find all your NextProgenitor indices inside the datasets of this tree (153231757will be in bounds).In practice, then, you could use the
treeOffsets()function to get the file and tree number (the first and third returns, respectively). Then, you would probably want to manually load (with h5py) the datasets of thisTree{X}group, from the given file, and then follow the NextProgenitor links as you have above.Hi Dylan,
Regarding the mass evolution of subhalos in IllustrisTNG in a merger, I plotted the mass versus time for two interacting subhalos (which merge at snapshot 50), as shown in the attached figure.
According to Figure 3 in Rodriguez-Gomez+ (2015), as you mentioned in this thread, I notice some differences compared to my figure. I wonder if the sharp drops in the subhalo mass history indicate other merging events occurring at those times. Additionally, why does the mass recover immediately after such rises or drops?
Thank you for your assistance.
This is called "switching", and can easily occur during mergers.
The masses recover because they have never actually physically decreased, simply Subfind decided to assign different particles to that subhalo at that particular snapshot.
So is it reasonable to take the mass at t_max (the time when the second progenitor reaches its peak mass) when there is a dip in the first progenitor at that time (according to my plot)?
I think you can see, by eye, reasonable values of the merger mass ratio, given the two lines you have plotted. That is, if you "unswitch" them by eye.
It would be best to come up with a definition that recovers such a value.
I do not see how the
numMergersfunction in Sublink deals with "switching." Does this mean it may over/under-estimate some merger ratios?That is true. It would be hard to get a perfect answer in all cases, as some trees are difficult. Averaging over a sizeable sample, however, would make this effect unimportant.